This protocol explains how to use the online integrated pipeline ‘peak-motifs‘ to predict motifs and binding sites in full-size peak sets obtained by chromatin 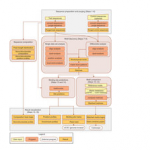 immunoprecipitation-sequencing (ChIP-seq) or related technologies. The workflow combines four time- and memory-efficient motif discovery algorithms to extract significant motifs from the sequences. Discovered motifs are compared with databases of known motifs to identify potentially bound transcription factors. Sequences are scanned to predict transcription factor binding sites and analyze their enrichment and positional distribution relative to peak centers. Peaks and binding sites are exported as BED tracks that can be uploaded into the University of California Santa Cruz (UCSC) genome browser for visualization in the genomic context. The protocol is illustrated with the analysis of a set of 6,000 peaks (8 Mb in total) bound by the Drosophila transcription factor Krüppel. The complete workflow is achieved in about 25 min of computational time on the Regulatory Sequence Analysis Tools (RSAT) Web server. This protocol can be followed in about 1 h.
immunoprecipitation-sequencing (ChIP-seq) or related technologies. The workflow combines four time- and memory-efficient motif discovery algorithms to extract significant motifs from the sequences. Discovered motifs are compared with databases of known motifs to identify potentially bound transcription factors. Sequences are scanned to predict transcription factor binding sites and analyze their enrichment and positional distribution relative to peak centers. Peaks and binding sites are exported as BED tracks that can be uploaded into the University of California Santa Cruz (UCSC) genome browser for visualization in the genomic context. The protocol is illustrated with the analysis of a set of 6,000 peaks (8 Mb in total) bound by the Drosophila transcription factor Krüppel. The complete workflow is achieved in about 25 min of computational time on the Regulatory Sequence Analysis Tools (RSAT) Web server. This protocol can be followed in about 1 h.
Incoming search terms:
- drosophila chip-seq analysis pipeline
- rna-seq analysis workflow
- whole exome sequencing analysis pipeline
- pgm ion torrent drosophila
- peak prediction chip seq
- ion torrent chip-seq drosophila
- how to use rtracklayer package for chip-seq data visualization
- drosophila chip-seq protocol
- diffsplice protocol
- cloud computing chip-seq
