List/Grid Chip-Seq
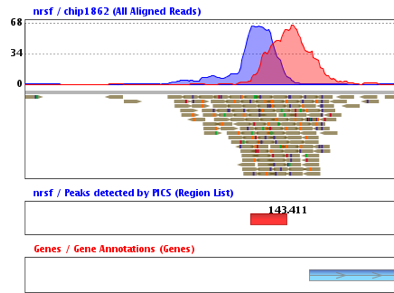
Biocondutor Packages for Analyzing Chip-seq Data
BayesPeak - Bayesian Analysis of ChIP-seq Data, This package is an implementation of the BayesPeak algorithm for peak-calling in ChIP-seq data.
Incoming search terms:Peakseqchip-seq ...
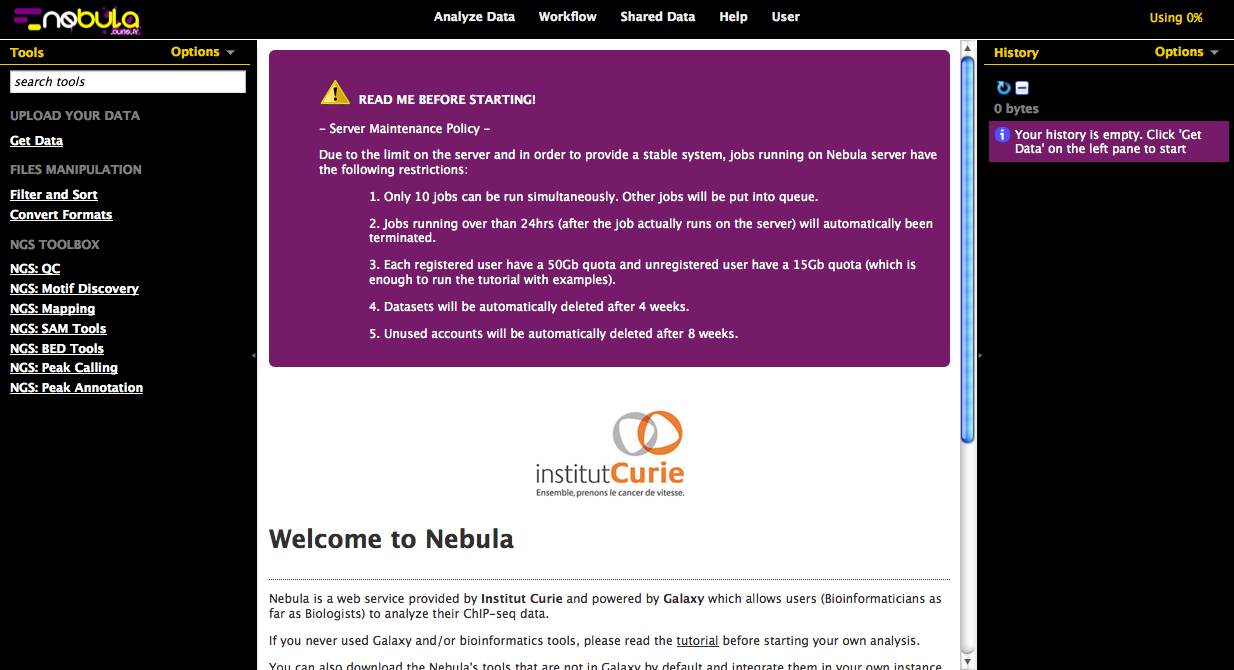
Nebula – a web-server for advanced ChIP-seq data analysis.
Nebula is for both bioinformaticians and biologists. It is based on the Galaxy open source framework. Galaxy already
Incoming search terms:chip-seq analysis pipelinechip seq analysis ...
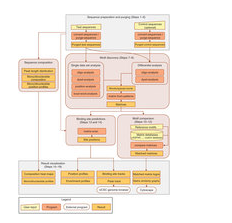
A complete workflow for the analysis of full-size ChIP-seq data sets using peak-motifs.
This protocol explains how to use the online integrated pipeline ‘peak-motifs‘ to predict motifs and binding sites in full-size peak sets obtained by chromatin
Incoming ...
GEM- Genome wide Event finding and Motif discovery
GEM is a software tool to study protein-DNA interaction using ChIP-Seq/ChIP-exo data. GEM resolves ChIP data into
Incoming search terms:gem chip-exoGemchip java Jarngs motif discovery ...
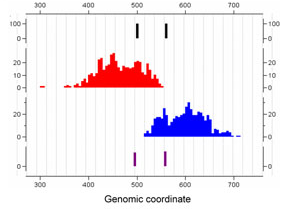
SeqSite: ChIP-Seq Binding Site Identification
SeqSite is an efficient and easy-to-use software tool implementing a novel method for identifying and pinpointing
Incoming search terms:seqsiteseqsigeseqeiteSeqSite ChIPSeqhow to use ...
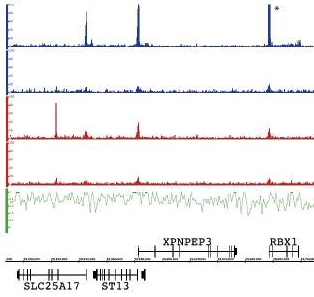
PeakSeq: a program for identifying and ranking peak regions in ChIP-Seq
PeakSeq is a program developed by Mark Gerstein’ lab for identifying and ranking peak regions in ChIP-Seq experiments. It takes as input, mapped reads
Incoming search terms:illumina ...
